Researchers have long been fascinated with the complex risk factors that lead to dementia in certain individuals. Known risk factors include a specific genetic mutation, called the Apolipoprotein E (APOE) ε4 allele. Roughly a quarter of the general population and 50 percent of Alzheimer’s disease patients have it. A recent study out of the University of Kentucky surprised researchers to find that even dementia patients without the mutation had strongly enriched ApoE proteins.
Researchers say the finding means the misfolded protein is considered a new biomarker for dementia.
“Dementia is very complex, but you can simplify it: the disease is caused by ‘gloppy proteins’ in the brain,” says lead investigator Dr. Peter T. Nelson, of the university’s Sanders-Brown Center on Aging and Department of Pathology, in a statement. “I’m not making light of it — these ‘sticky’ misfolded proteins often end up destroying the brain, the mind, the memories and everything else for millions of people who suffer from dementia. We want to understand specifically which proteins are the problem.”
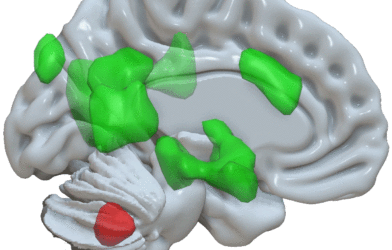
The researchers examined 40 participants, some of them healthy, some of them in different stages of dementia. They used mass spectronomy, a detection technique used to obtain information about proteins, to define the complete set of proteins (proteome) from the participants’ amygdalae. The amygdala is the part of the brain that processes emotions and memories associated with fear. It is often affected from the very beginning of dementia.
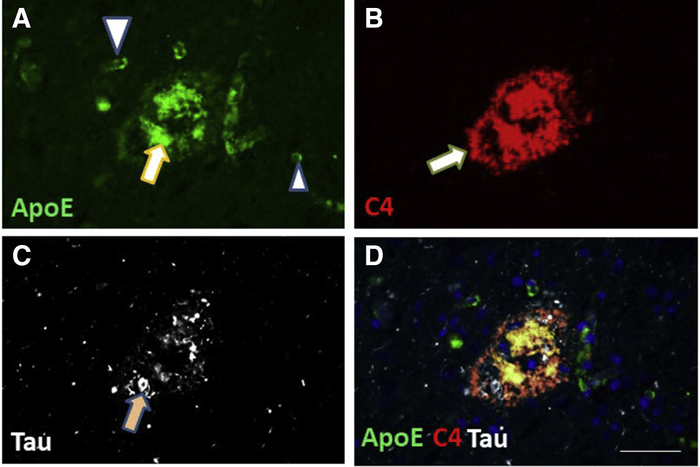
As expected, the researchers found proteins associated with dementia in the brains of the dementia patients, such as Tau, Aβ, and α-Synuclein. They found Tau and Aβ proteins in some of the “healthy” brains as well. This was also expected by the researchers. But their data further revealed that “ApoE was found to be aberrantly aggregated in dementia brains,” making it a stronger indicator, or biomarker, of dementia than Tau, Aβ, or α-Synuclein, according to the original study.
“ApoE peptides were strongly enriched in dementia cases, including from individuals lacking the APOE ε4 genotype,” the researchers found. Thus, showing “that the ApoE protein may play active disease-driving mechanistic roles in persons lacking the APOE ε4 allele,” the research states.
“Our study adds to an evolving appreciation of multiple misfolded proteins in the human brain and moves the field forward by emphasizing that ApoE may be a strong contributor to the dementia prototype, even in individuals who do not have the disease-driving version of the APOE gene,” explained Dr. Nelson. “Even in persons lacking the APOE ε4 allele, ApoE may indeed be among the most impactful ‘gloppy proteins’ in aging brains.”
The research was the first study of this size including a control group. Its findings were published in The American Journal of Pathology.